The functional specialization of visual cortex emerges from training parallel pathways with self-supervised predictive learning
Shahab Bakhtiari, Patrick Mineault, Timothy Lillicrap, Christopher Pack, Blake Richards
McGill University, Mila, DeepMind
NeurIPS 2021 (spotlight presentation, <3% selected)
Code | Preprint
Abstract
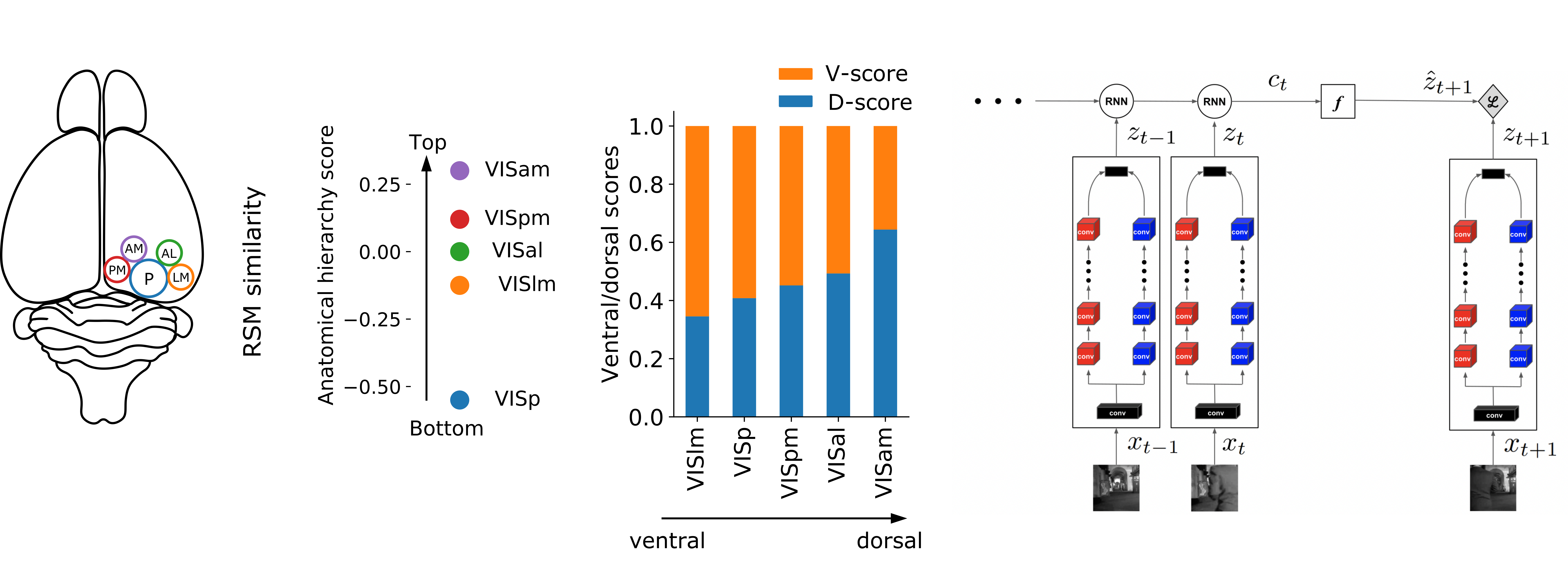
The visual system of mammals is comprised of parallel, hierarchical specialized pathways. Different pathways are specialized in so far as they use representations that are more suitable for supporting specific downstream behaviours. In particular, the clearest example is the specialization of the ventral ("what") and dorsal ("where") pathways of the visual cortex. These two pathways support behaviours related to visual recognition and movement, respectively. To-date, deep neural networks have mostly been used as models of the ventral, recognition pathway. However, it is unknown whether both pathways can be modelled with a single deep ANN. Here, we ask whether a single model with a single loss function can capture the properties of both the ventral and the dorsal pathways. We explore this question using data from mice, who like other mammals, have specialized pathways that appear to support recognition and movement behaviours. We show that when we train a deep neural network architecture with two parallel pathways using a self-supervised predictive loss function, we can outperform other models in fitting mouse visual cortex. Moreover, we can model both the dorsal and ventral pathways. These results demonstrate that a self-supervised predictive learning approach applied to parallel pathway architectures can account for some of the functional specialization seen in mammalian visual systems.
Prefered stimuli
For a qualitative understanding of the difference between the two pathways of our model, we have created optimal stimuli for each pathway, using gradient ascent on the input space to maximize the layer activations. The stimuli are created by maximizing the activation of the layers with the highest similarity with VISlm (for the ventral-like pathway) and VISam (for the dorsal-like pathway). The optimal stimuli for the ventral-like area mostly contain static textures and patterns with some local moving components, while for the dorsal-like area, the optimal stimuli mostly contain distributed motion. Samples of the optimal stimuli can be found here.
Acknowledgements
We thank Iris Jianghong Shi, Michael Buice, Stefan Mihalas, Eric Shea-Brown, and Bryan Tripp for helpful discussions. We also thank Pouya Bashivan and Alex Hernandez-Garcia for their suggestions on the manuscript. This work was supported by a NSERC (Discovery Grant: RGPIN-2020-05105; Discovery Accelerator Supplement: RGPAS-2020-00031), Healthy Brains, Healthy Lives (New Investigator Award: 2b-NISU-8; Innovative Ideas Grant: 1c-II-15), and CIFAR (Canada AI Chair; Learning in Machine and Brains Fellowship). CCP was funded by a CIHR grant (MOP-115178). This work was also funded by the Canada First Research Excellence Fund (CFREF Competition 2, 2015-2016) awarded to the Healthy Brains, Healthy Lives initiative at McGill University, through the Helmholtz International BigBrain Analytics and Learning Laboratory (HIBALL).